Bayesian analysis for small molecule sensors
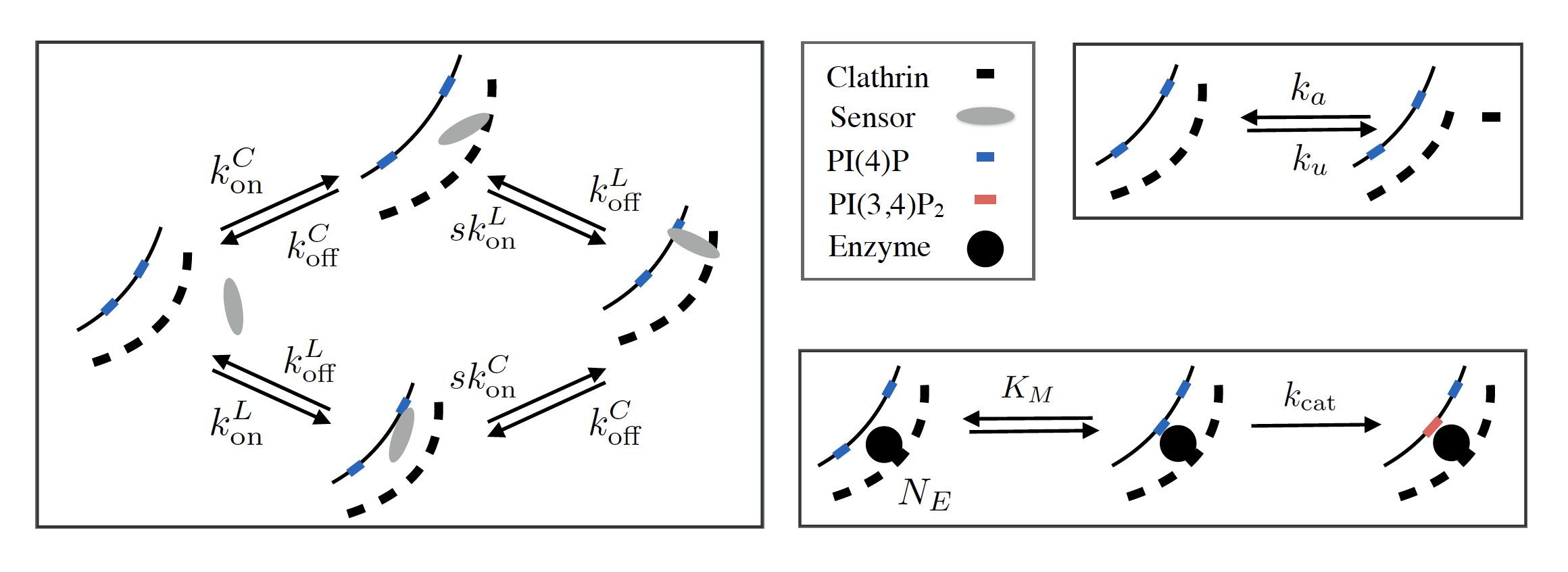
The combination of gene editing and lattice light-sheet microscopy has opened a new era in cell biology. It is now possible, in principle, to fluorescently label any biologically relevant molecule and measure its dynamics across the whole cell in real time. Not only can proteins be labeled, by directly appending the code for a fluorescent domain to the gene of interest, but so can small molecules and ions, by hijacking the naturally occurring proteins that transduce information about their concentrations, using them to create artificial fluorescent probes. Stitching these sensors to other protein domains that specifically bind to different parts of the cell can provide additional spatial resolution, detecting even a small number of copies of the molecule of interest in the vicinity of a certain target structure. A number of small molecules such as cyclic AMP and phosophotidylinositol (PI) play crucial roles in signal transduction, and a complete understanding how the signal is processed requires careful measurements of their spatiotemporal dynamics in response to specific stimuli.
Inferring the dynamics of the molecules of interest from the observed behavior of the fluorescent probes, however, presents new challenges. The timescale of concentration changes for the target molecule is often comparable to the timescale for the probe to bind it, requiring a detailed kinetic model to back out the desired information. While it is usually straightforward to write down the kinetics for a well-characterized probe, the multi-domain molecules developed for the latest high-resolution measurements require a large number of unknown kinetic parameters, whose in vitro values do not necessarily reflect their behavior in the cytoplasm. Using these high-dimensional models correctly requires great care, because they easily give rise to “sloppy” directions in parameter space, which are unconstrained by the available data. I have developed some expertise in using Bayesian analysis to guide careful interpretation of the results of this kind of experiment, in the context of detecting phosphorylated PI molecules recruited to sites of clathrin-mediated endocytosis (Marsland III, 2017; He et al., 2017).
References
- Marsland III, R. A. (2017). The edge of thermodynamics: driven steady states in physics and biology [PhD thesis]. Massachusetts Institute of Technology.
- He, K., Marsland III, R., Upadhyayula, S., Song, E., Dang, S., Capraro, B. R., Wang, W., Skillern, W., Gaudin, R., Ma, M., & others. (2017). Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature, 552(7685), 410.