Tools for quantitative prediction in microbial communities
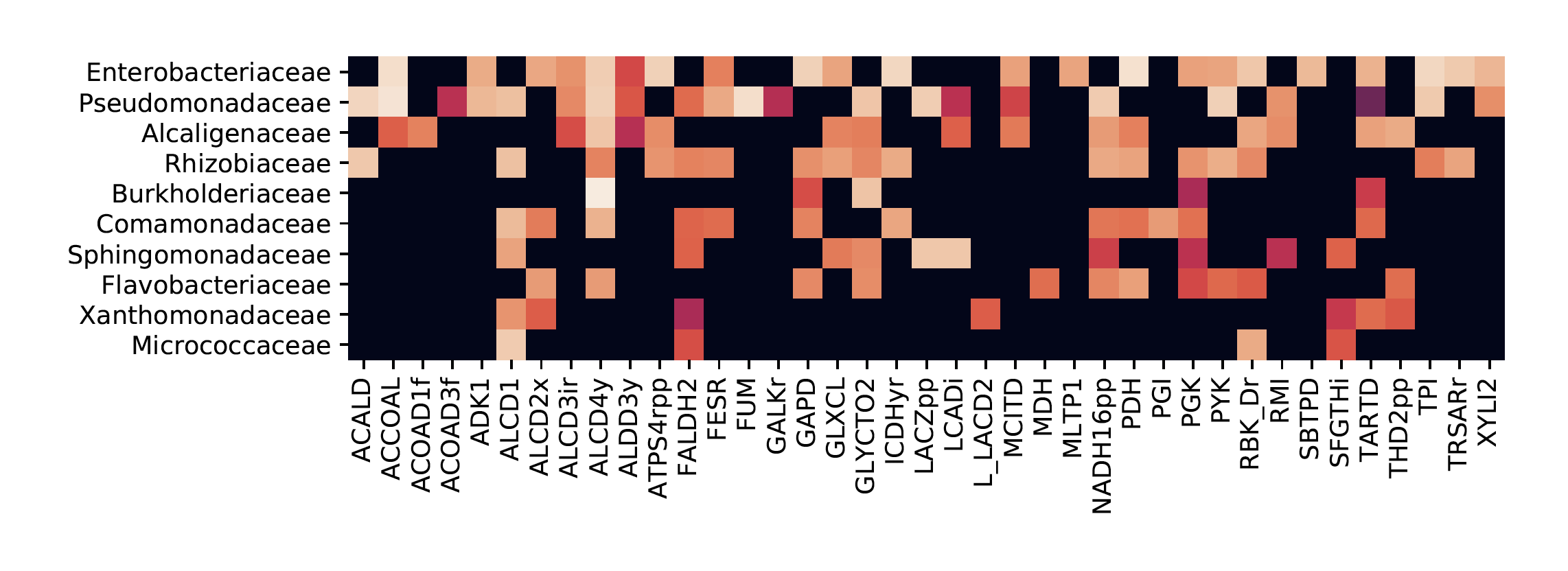
The concept of a “species” is notoriously ill-defined in the microbial realm. Instead of the sharp distinctions between different kinds of plants and animals we find in the macroscopic world, microbes manifest a fuzzier continuum of phenotypes. Given this blurriness, it should come as no surprise that it is difficult to predict “species”-level composition of microbial communities. But natural microbial communities do have sufficiently stable functional properties to be effectively utilized by macroscopic organisms to accomplish definite tasks. This means that there should be some community-level quantities that are reproducible and controllable, and some “relevant variables” that reliably control them, but what these quantities could be remains an open question.
I have recently been following up on an observation by some of my colleagues that the composition of communities grown on single carbon sources is reproducible at a sufficiently high taxonomic level. Specifically, the family-level structure of the communities is sensitive to the choice of carbon source, but is strongly conserved for different communities, sampled from different field sites, when grown on the same carbon source. My first goal was to identify the chemical features of the carbon source that determine the community structure, using standard machine learning algorithms to predict community composition on novel carbon sources based on chemical similarity, and comparing these predictions with the measurements performed by my collaborators.
The next step would be to develop a method for inferring reproducible observables directly from the data, instead of using the historically contingent family groupings (which turn out not to generate reproducible measurements in other experimental settings). An interesting theoretical procedure for this was proposed some time ago, using the eigenvectors of an individual-level, environment-specific interaction matrix. It is currently impossible to measure the entries of this matrix directly, and so practical implementation of this program requires a controlled procedure for estimating the matrix from the data. I have made some progress towards developing such a procedure, applying it to my collaborators’ data, and exploring other ways of obtaining meaningful functional groupings from the inferred matrix.