Statistical physics approaches to community ecology
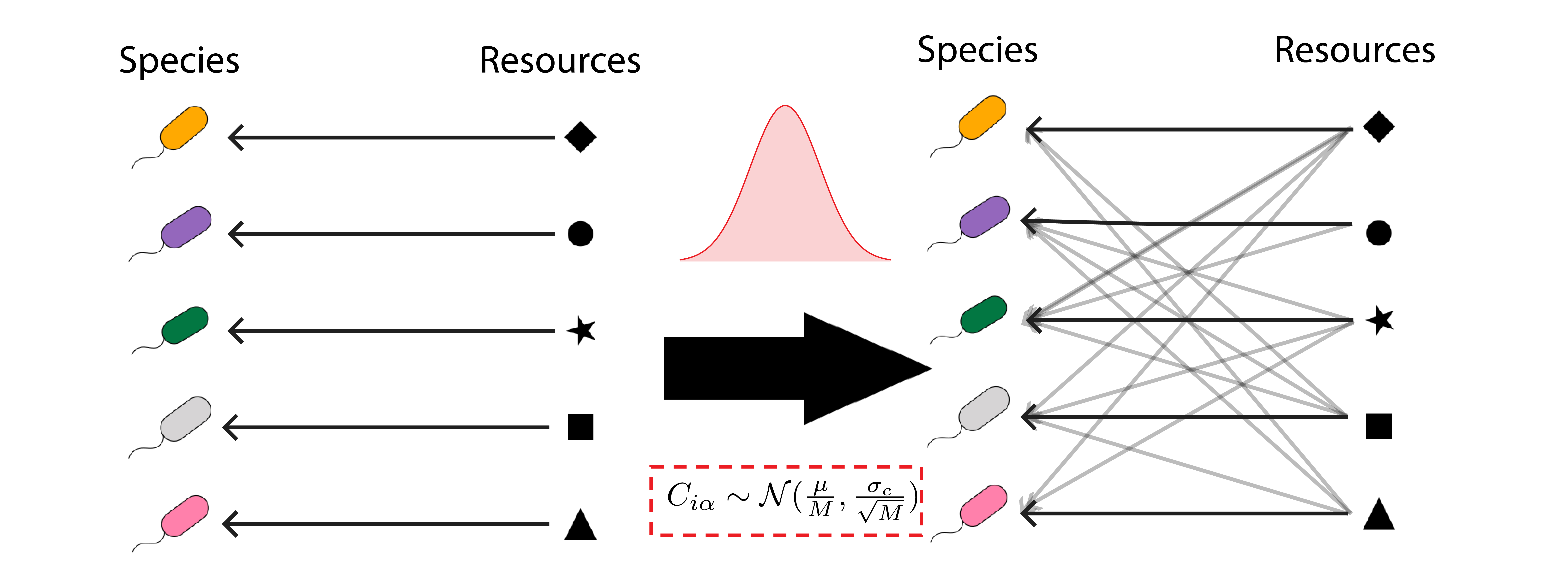
Microbial ecosystems manifest one of the main technical obstacles to predictive modeling of biological systems, which arises from their extreme heterogeneity. The standard methods of statistical mechanics produce powerful and robust predictions about the behavior of large collections of functionally identical particles, which are easily extended to mixtures of a few different kinds of components. But in most biological systems, the number of distinct types of components is unmanageably large, whether we are considering the 20,000 different genes in the human genome, or the 20,000 distinct metabolites involved in biochemical networks. Microbial ecosystems reproduce this same difficulty on a bigger length scale, with hundreds to thousands of coexisting “species” in a given natural community. The development of inexpensive DNA sequencing has made these systems an attractive setting for developing and testing new theoretical methods for overcoming this hurdle. Highly diverse microbial communities can be readily found on any plant leaf, pond droplet, or grain of soil, and sequencing technology makes it possible to determine which species are present there and in what abundance.
My colleagues and I have been adapting methods from the physics of disordered materials to this new setting, deriving predictions about the diversity, composition and sensitivity of heterogeneous communities based on summary statistics about consumption preferences and byproduct secretion in the regional species pool. The key assumption behind these methods is that the parameters characterizing each species can be modeled as independent random variables drawn from a given probability distribution. This seems like an extreme assumption, given how much specific structure is already known about microbial behavior. But we have shown that at sufficiently high levels of diversity, the strong interactions between specific pairs of organisms based on known mechanisms can be overwhelmed by the sheer number of weak, uncharacterized interactions with all the other species in the system (Cui et al., 2021).
To check our calculations and facilitate hypothesis generation, I have also been developing a Python package for rapidly simulating diverse microbial ecosystems with randomly sampled parameters (Marsland III et al., 2020). Using this package, I have shown that the robust patterns observed in large-scale surveys of microbial diversity (such as the Human Microbiome Project and Earth Microbiome Project) reflect the “typical” structure of diverse communities (Marsland III et al., 2020).
I have also used this software to study some novel ecological phenomena that can arise in self-sufficient microbial ecosystems (Marsland III et al., 2019). Since microbes generically produce usable byproducts whenever they consume an energy source, they can autonomously generate a complex chemical environment that supports a diverse community even when only a single energy source is supplied externally. This differs from the standard case considered in macroscopic ecology, where environmental conditions are either fixed or modified only through consumption. By performing simulations of this extreme case of a fully self-generated environment, I identified a transition in community structure as a function of the supply rate of the externally provided resource. At high supply rates, the community-level metabolic network percolates, and all possible byproducts are produced at sufficiently high concentrations to significantly contribute to the growth of some set of species. The scaling of diversity with the number of supplied resources in this regime agrees with analytic predictions obtained for “typical” communities. But at low supply rates, only species that directly consume the externally supplied resource can survive, and the resulting low-diversity state no longer satisfies the assumptions for calculating “typical” community properties.
References
- Cui, W., Marsland III, R., & Mehta, P. (2021). Diverse communities behave like typical random ecosystems. Physical Review E, 104, 034416.
- Marsland III, R., Cui, W., Goldford, J., & Mehta, P. (2020). The Community Simulator: A Python package for microbial ecology. PLoS ONE, 15, e0230430.
- Marsland III, R., Cui, W., & Mehta, P. (2020). A minimal model for microbial biodiversity can reproduce experimentally observed ecological patterns. Scientific Reports, 10, 3308.
- Marsland III, R., Cui, W., Goldford, J., Sanchez, A., Korolev, K., & Mehta, P. (2019). Available energy fluxes drive a transition in the diversity, stability, and functional structure of microbial communities. PLoS Computational Biology, 15(2), e1006793.